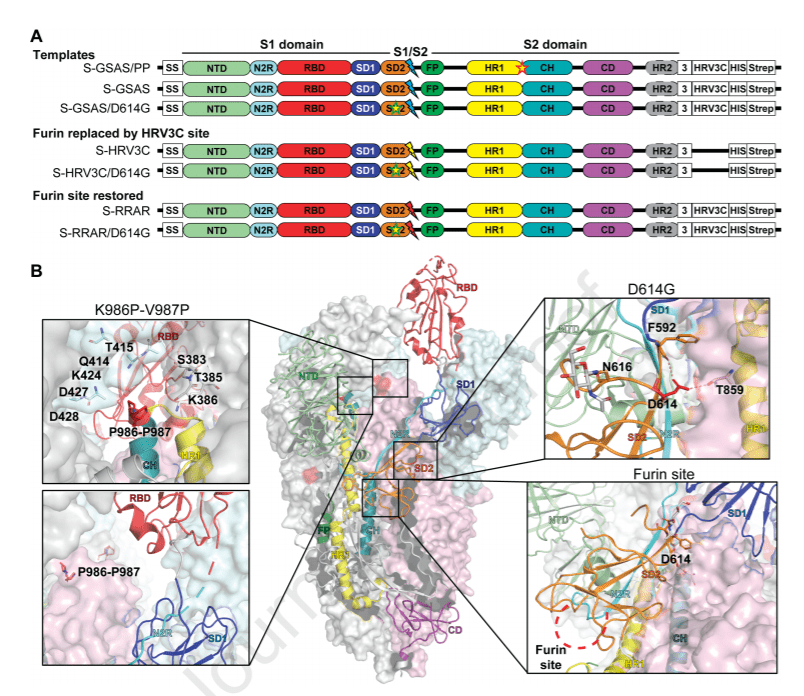
The SARS-CoV-2 spike (S) protein is the target of vaccine design efforts to end the COVID-19 pandemic. Despite a low mutation rate, isolates with the D614G substitution in the S protein appeared early during the pandemic, and are now the dominant form worldwide.
In this paper are explored spike conformational changes and the effects of the D614G mutation on a soluble S ectodomain construct. Cryo-EM structures reveal altered RBD disposition; antigenicity and proteolysis experiments reveal structural changes and enhanced furin cleavage efficiency of the G614 variant.
Furthermore, furin cleavage alters the up/down ratio of the Receptor Binding Domains (RBD) in the G614 S ectodomain, demonstrating an allosteric effect on RBD positioning triggered by changes in the SD2 region, that harbors residue 614 and the furin cleavage site.
This paper results elucidate SARS-CoV-2 spike conformational landscape and allostery, and have implications for vaccine design.

Leave a Reply